Recently, COVID-19 has raised significant public health concerns globally. However, the some pivotal structure of the causative agent, SARS-CoV-2, is yet not quite clear. In this study, Dan Su et al. determined the structure of SARS-CoV-2 nonstructural protein 9 (nsp9), an RNA-binding protein that is essential for CoV replication, which may be essential for the regulation of viral RNA replication and transcription.

Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the causative agent of a potentially fatal disease named coronavirus disease 2019 (COVID-19), has raised significant public health concerns globally. To date, the COVID-19 pandemic has caused millions of people to be infected with SARS-CoV-2 worldwide.
It has been known since the 2003 SARS epidemic that coronaviruses (CoVs) have large RNA genomes, the replication of which requires an RNA-dependent RNA replication/transcription complex. CoV nonstructural proteins (Nsps) play pivotal roles in the assembly of this complex and associated enzymatic functions in virus genomic replication. Several smaller nonenzymatic Nsps assist with RNA-dependent RNA polymerase function. In this study, Dan Su et al. determined the structure of SARS-CoV-2 nonstructural protein 9 (nsp9), an RNA-binding protein that is essential for CoV replication. Its homotetrameric structure with two stable dimeric interfaces provids a structural basis for understanding the mechanisms of RNA-binding protein self-assembly, which may be essential for the regulation of viral RNA replication and transcription.
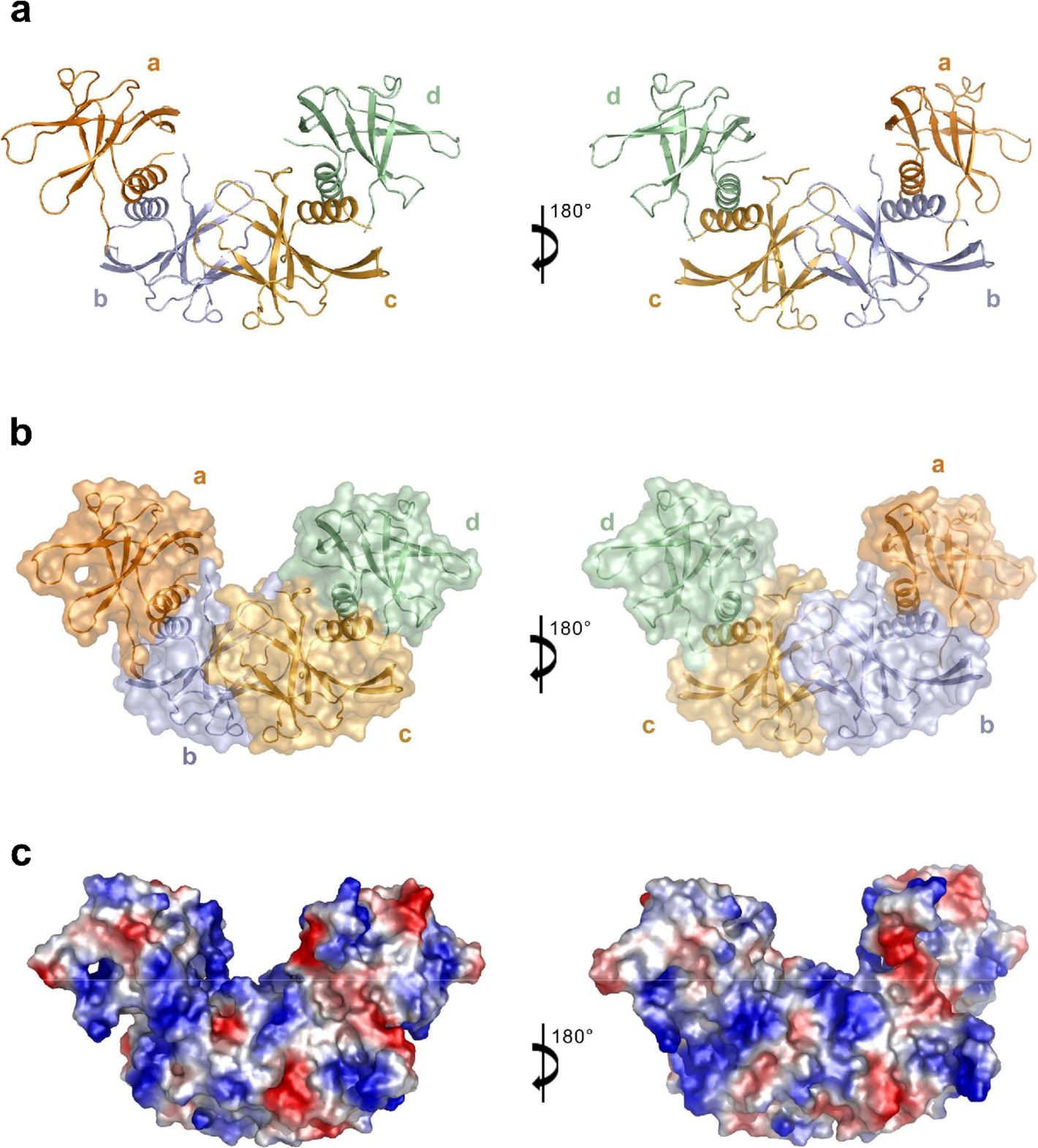
Fig. 1 The Structure of the SARS-CoV-2 nsp9 tetramer
Article Access: https://https://link.springer.com/article/10.1186/s43556-020-00005-0
Website for Molecular Biomedicine: https://www.springer.com/journal/43556
Looking forward to your contributions