The spread of coronavirus disease 2019 (COVID‐19) has become a global pandemic and public health problem. Although unprecedented efforts had been concentrated to identify and isolate individuals with risk of SARS‐CoV‐2 infection, clinicians are facing tremendous difficulties in efficiently and quickly diagnosing these patients due to massive volume of suspected cases. Herein, Prof. Minghua Zheng et al developed a machine‐learning algorithm based on radiomics to assist clinical diagnosis.
Clinicians have been faced with the challenge of differentiating between severe acute respiratory syndrome associated coronavirus 2 (SARS‐CoV‐2) infected pneumonia (NCP) and influenza A infected pneumonia (IAP), a seasonal disease that coincided with the outbreak. Thus, the authors aim to develop a machine‐learning algorithm based on radiomics to distinguish NCP from IAP by texture analysis based on computed tomography (CT) imaging (Fig. 1).
Forty‐one NCP and 37 IAP patients admitted from January to February 6, 2019 admitted to two hospitals in Wenzhou, China. All patients had undergone chest CT examination and blood routine tests prior to receiving medical treatment. NCP was diagnosed by real‐time RT‐PCR assays. Eight of 56 radiomic features extracted by LIFEx were selected by least absolute shrinkage and selection operator regression to develop a radiomics score and subsequently constructed into a nomogram to predict NCP with area under the operating characteristics curve of 0.87 (95% confidence interval: 0.77‐0.93). The nomogram also showed excellent calibration with Hosmer‐Lemeshow test yielding a nonsignificant statistic (P = .904). The novel nomogram may efficiently distinguish between NCP and IAP patients. The nomogram may be incorporated to existing diagnostic algorithm to effectively stratify suspected patients for SARS‐CoV‐2 pneumonia.
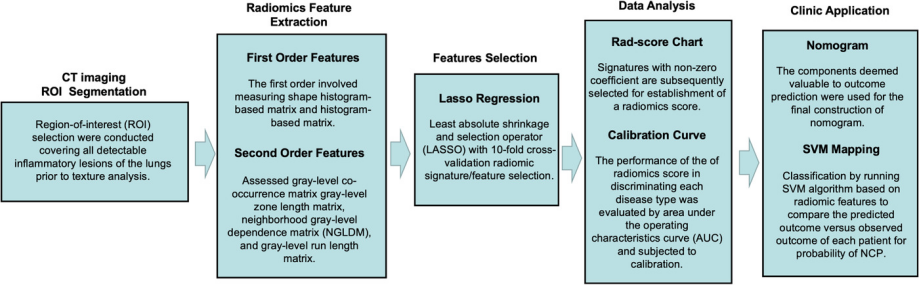
Fig. 1 Radiomics‐based machine learning workflow
Article Access: https://onlinelibrary.wiley.com/doi/10.1002/mco2.14
Website for MedComm: https://onlinelibrary.wiley.com/journal/26882663
Looking forward to your contributions.